Currently part of the VIB Bioimaging Core Gent Team.
Interested in ❄️🔬 #CryoEM and #Cytokines.

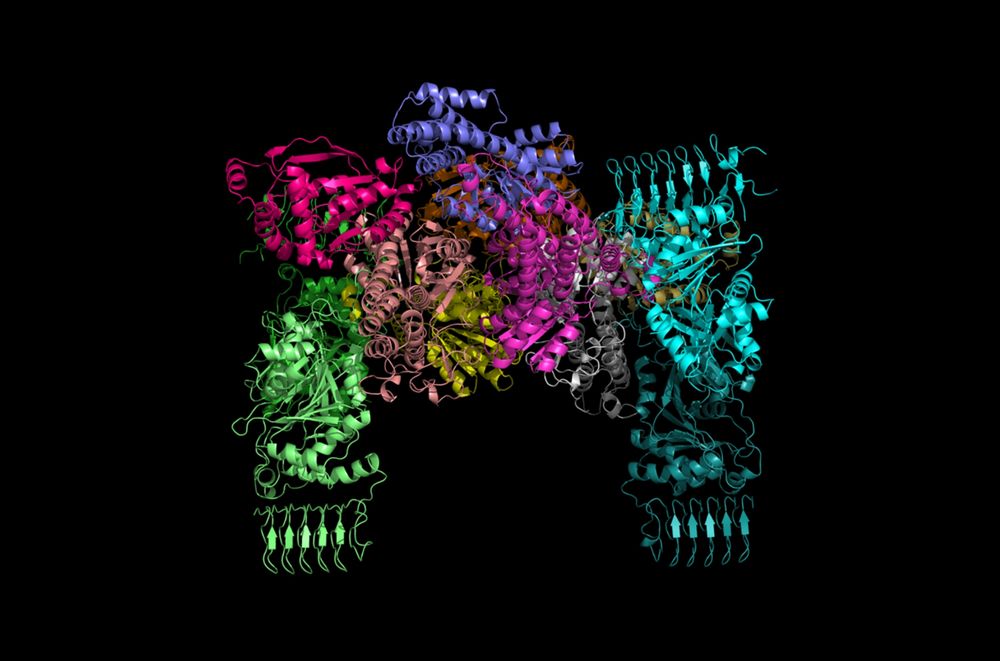
Fantastic collaboration & massive effort from Sophie Lucas Lab @ de Duve Institute. #CryoEM with @savvideslab.bsky.social.
Experiment-guided AlphaFold3 resolves accurate protein ensembles.
doi.org/10.1101/2025...
AlphaFold3 is incredible, but has crucial limitations: it typically collapses to a single conformation, ignoring the inherent dynamics of proteins. And it can be wrong. Here's a solution. 🧵👇

Experiment-guided AlphaFold3 resolves accurate protein ensembles.
doi.org/10.1101/2025...
AlphaFold3 is incredible, but has crucial limitations: it typically collapses to a single conformation, ignoring the inherent dynamics of proteins. And it can be wrong. Here's a solution. 🧵👇
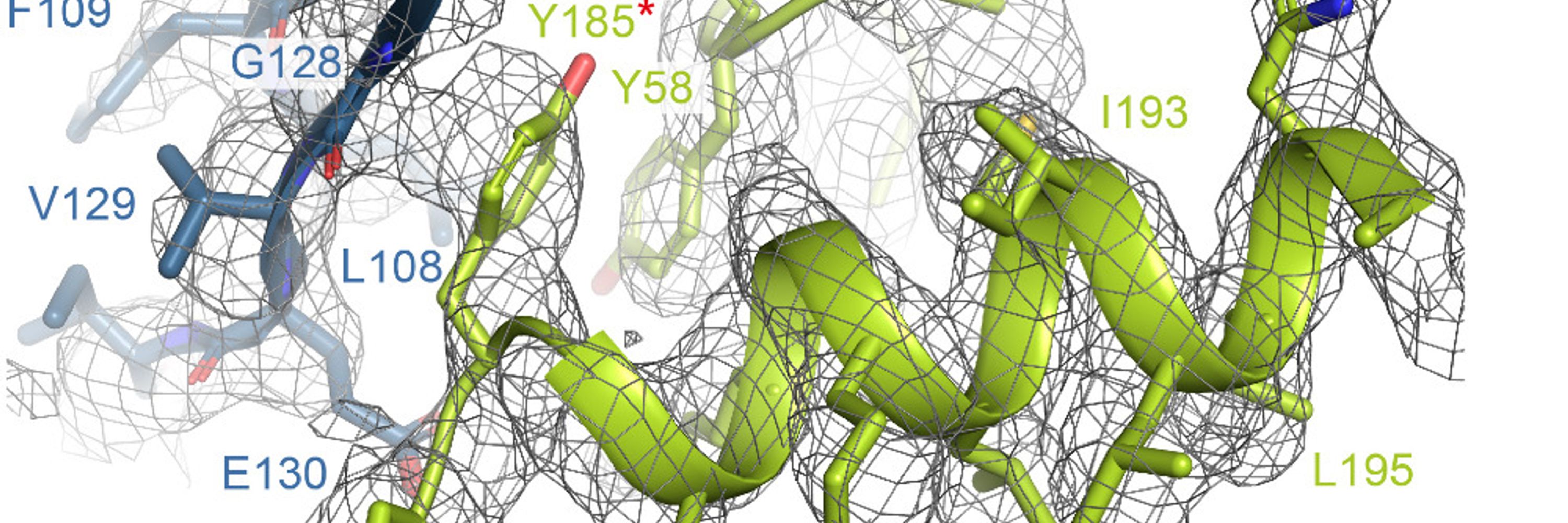
A 1-class ab initio in relion, then local refinement in relion (1.8deg searches+blush) gives a nominally 3.3Å map; 3.5 Å w/out blush.
A 1-class ab initio in relion, then local refinement in relion (1.8deg searches+blush) gives a nominally 3.3Å map; 3.5 Å w/out blush.
edu.nl/pvhxv
Please try it out. LocScale is evolving & we are actively building in new features and improvements. Your feedback will help us shape the next steps.
Code: cryotud.github.io/locscale/
Work by @alok-bharadwaj.bsky.social and Reinier.
Taking two half maps as input, LcoScale-2.0 produces feature-enhanced maps along with a robust confidence score that guides objective map interpretation.
cryotud.github.io/locscale/
edu.nl/pvhxv
Please try it out. LocScale is evolving & we are actively building in new features and improvements. Your feedback will help us shape the next steps.
Code: cryotud.github.io/locscale/
Work by @alok-bharadwaj.bsky.social and Reinier.
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
Fantastic collaboration & massive effort from Sophie Lucas Lab @ de Duve Institute. #CryoEM with @savvideslab.bsky.social.

Fantastic collaboration & massive effort from Sophie Lucas Lab @ de Duve Institute. #CryoEM with @savvideslab.bsky.social.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
"Single-domain antibodies directed against hemagglutinin and neuraminidase protect against influenza B viruses".
Great collab between @savvideslab.bsky.social (we performed #CryoEM) and Xavier Saelens Lab, spearheaded by Arne Matthys.
www.nature.com/articles/s41...

"Single-domain antibodies directed against hemagglutinin and neuraminidase protect against influenza B viruses".
Great collab between @savvideslab.bsky.social (we performed #CryoEM) and Xavier Saelens Lab, spearheaded by Arne Matthys.
www.nature.com/articles/s41...
EHD2 forms rings on caveolae necks, in contrast to most EHDs forming helices. We determined its structure on membranes and show that N-term acts as a spacer
By Elena, Vasya @vasiliimikirtumov.bsky.social, Jeff &Oli Daumke www.biorxiv.org/content/10.1...
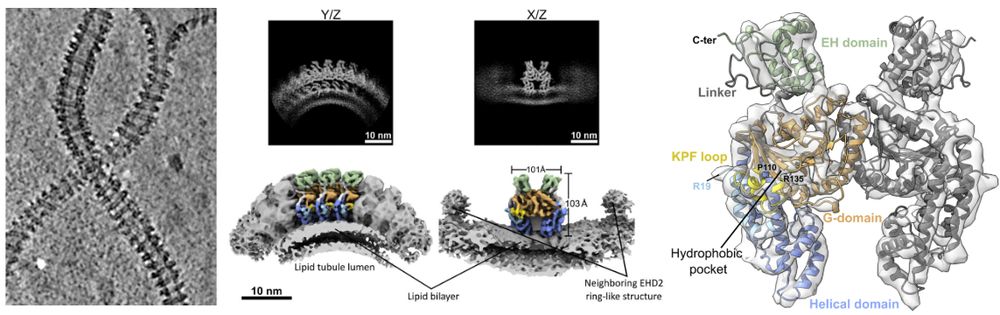
EHD2 forms rings on caveolae necks, in contrast to most EHDs forming helices. We determined its structure on membranes and show that N-term acts as a spacer
By Elena, Vasya @vasiliimikirtumov.bsky.social, Jeff &Oli Daumke www.biorxiv.org/content/10.1...
www.sciencedirect.com/science/arti...

www.sciencedirect.com/science/arti...
Taking two half maps as input, LcoScale-2.0 produces feature-enhanced maps along with a robust confidence score that guides objective map interpretation.
cryotud.github.io/locscale/
Taking two half maps as input, LcoScale-2.0 produces feature-enhanced maps along with a robust confidence score that guides objective map interpretation.
cryotud.github.io/locscale/
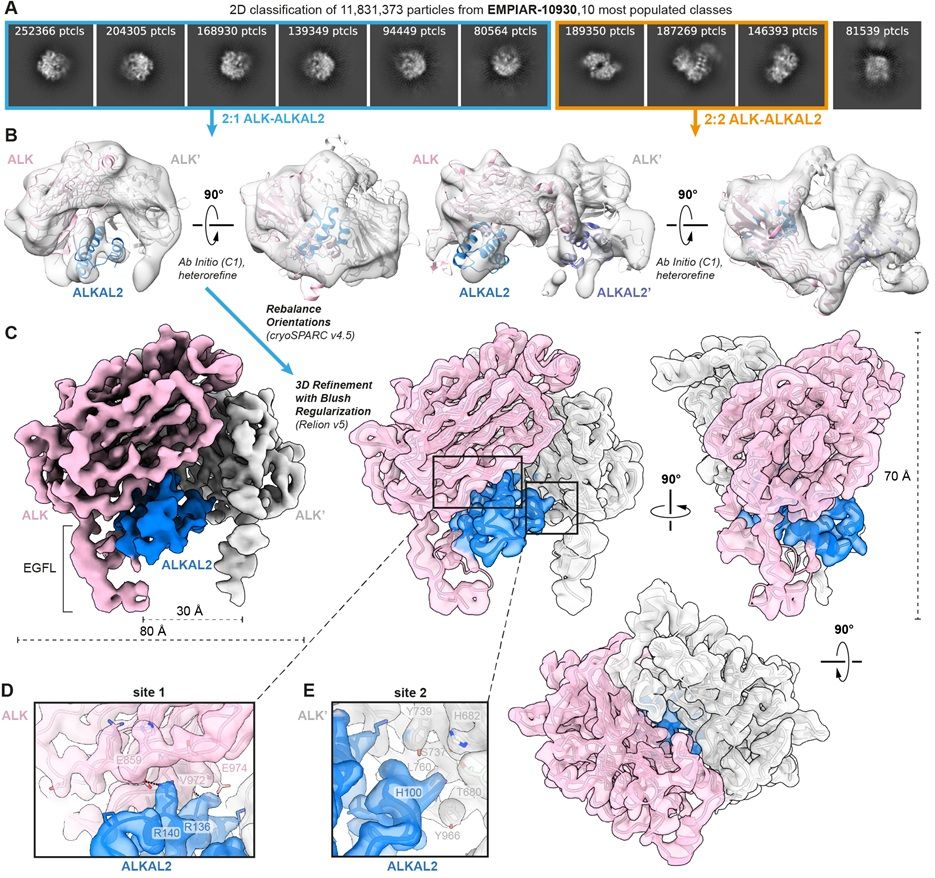
journals.plos.org/plosbiology/... ;
revealing the ALK-ALKAL2 assembly with 2:1 stoichiometry and resolving the conundrum introduced by the originally reported 2:2 complex. ALK/LTK-cytokine signalling getting clearer!

journals.plos.org/plosbiology/... ;
revealing the ALK-ALKAL2 assembly with 2:1 stoichiometry and resolving the conundrum introduced by the originally reported 2:2 complex. ALK/LTK-cytokine signalling getting clearer!
Featuring a new automatic Micrograph Junk Detector that labels and rejects contaminants, new tools for curating subsets of particles, performance and stability fixes, and more!
Full changelog: cryosparc.com/updates
#cryoEM
Featuring a new automatic Micrograph Junk Detector that labels and rejects contaminants, new tools for curating subsets of particles, performance and stability fixes, and more!
Full changelog: cryosparc.com/updates
#cryoEM
📖 www.science.org/doi/10.1126/...
But what changed compared to the original preprint?
Also, I feel i should post Movie 1 🎥, that inspired the cover. Back when I did the original bluesky thread, movies were not available.
📖 www.science.org/doi/10.1126/...
But what changed compared to the original preprint?
Also, I feel i should post Movie 1 🎥, that inspired the cover. Back when I did the original bluesky thread, movies were not available.
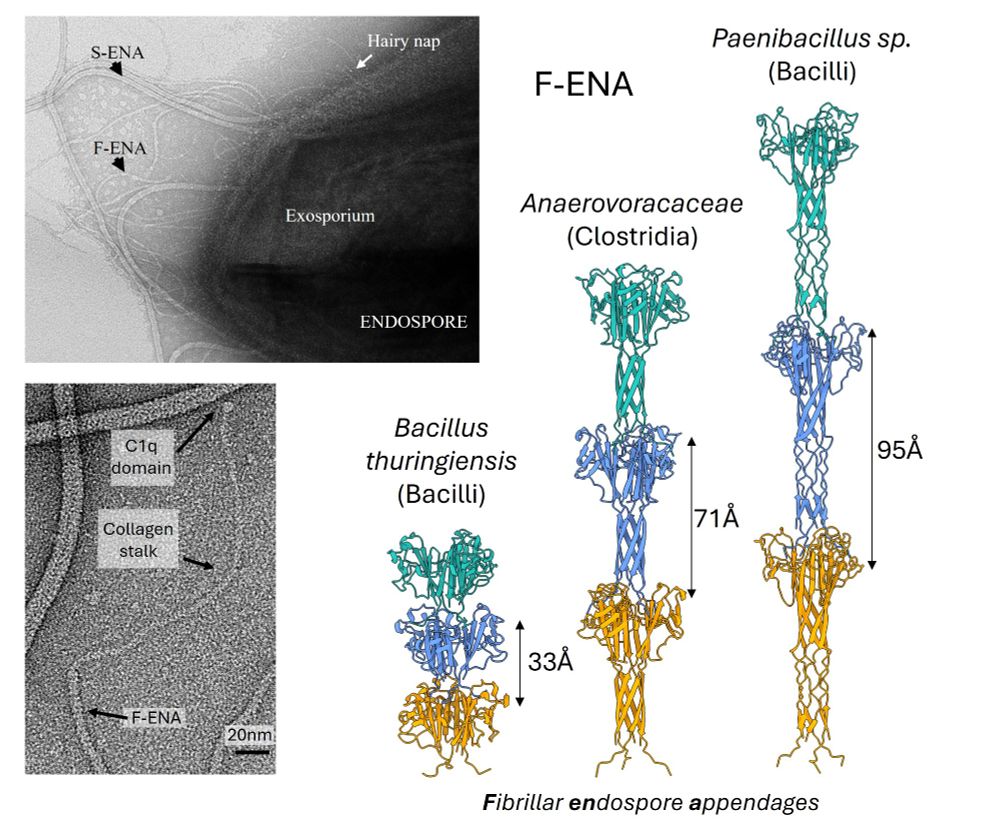
Yes, Micro ISOlation (MISO) sample preparation makes it possible.
www.biorxiv.org/content/10.1...
Happy to be part of this exciting story and wonderful collaboration with the Rouslan Efremov lab.
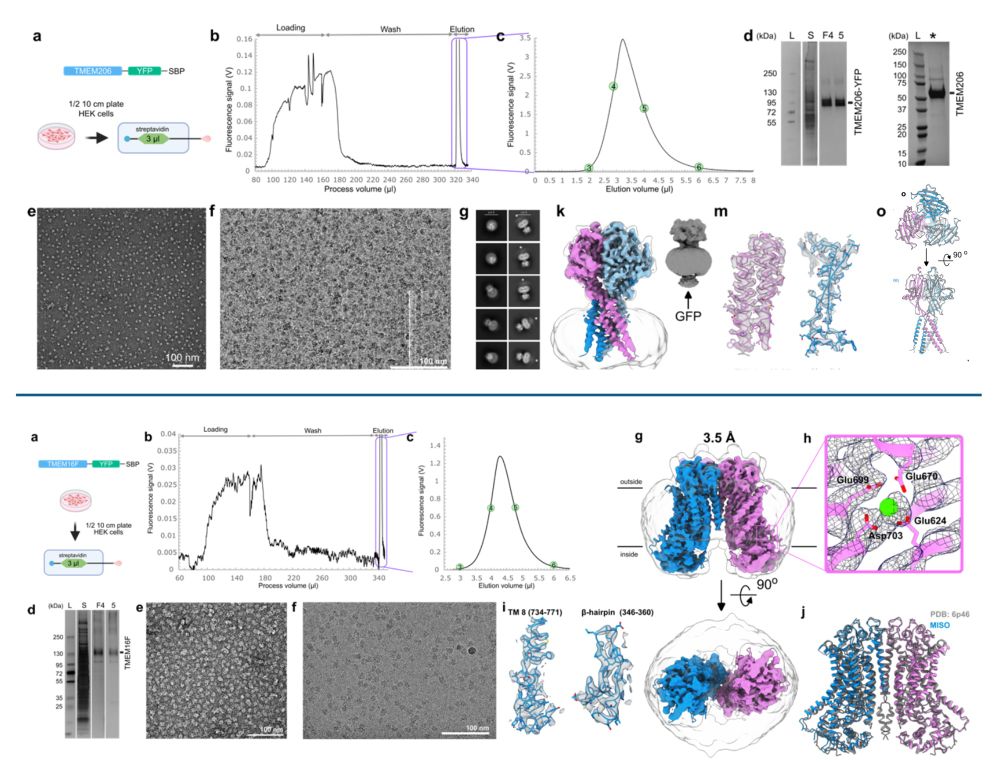
Yes, Micro ISOlation (MISO) sample preparation makes it possible.
www.biorxiv.org/content/10.1...
Happy to be part of this exciting story and wonderful collaboration with the Rouslan Efremov lab.

