Roberto Chica Lab
@chicalab.bsky.social
Our research group at the University of Ottawa specializes in computational enzyme design.
mysite.science.uottawa.ca/rchica/
mysite.science.uottawa.ca/rchica/
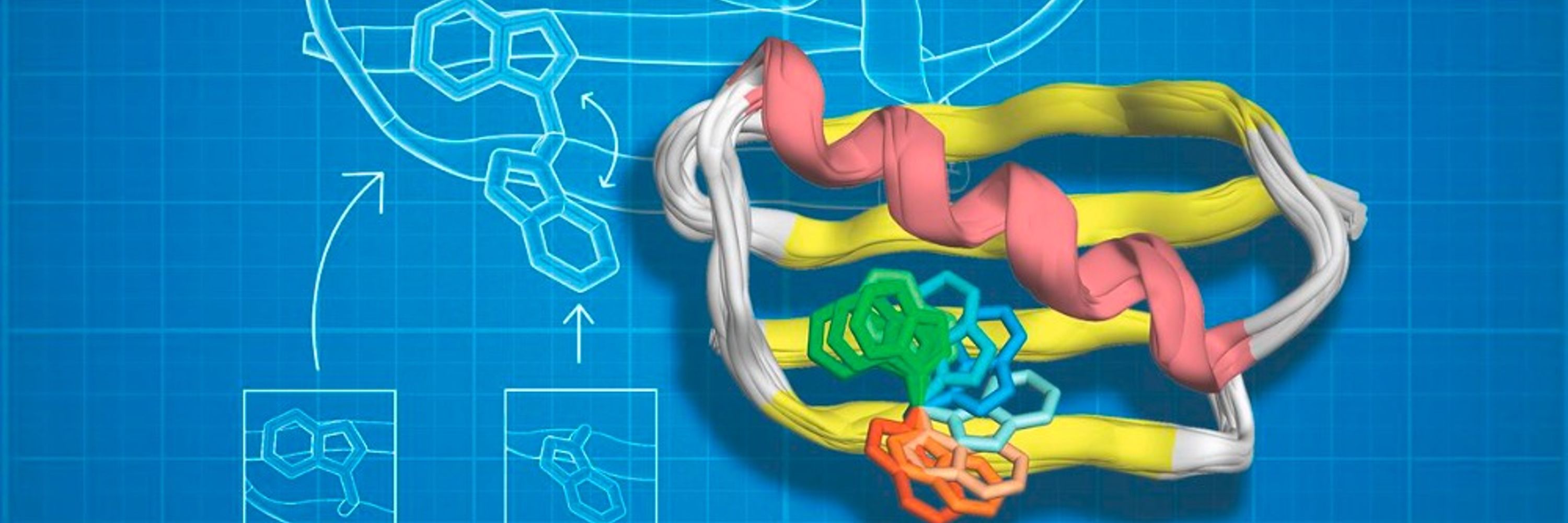
Thrilled to share that our latest article is now out in final form! A great collaboration with @fraserlab.com and @silviaosuna.bsky.social.
Distal mutations enhance catalysis in designed enzymes by facilitating substrate binding and product release
www.nature.com/articles/s41...
Distal mutations enhance catalysis in designed enzymes by facilitating substrate binding and product release
www.nature.com/articles/s41...

Distal mutations enhance catalysis in designed enzymes by facilitating substrate binding and product release - Nature Communications
Distal mutations, though far from the active site, enhance Kemp eliminase catalysis by tuning conformational dynamics that facilitate substrate binding and product release, thereby promoting the full catalytic cycle.
www.nature.com
September 30, 2025 at 3:42 PM
Thrilled to share that our latest article is now out in final form! A great collaboration with @fraserlab.com and @silviaosuna.bsky.social.
Distal mutations enhance catalysis in designed enzymes by facilitating substrate binding and product release
www.nature.com/articles/s41...
Distal mutations enhance catalysis in designed enzymes by facilitating substrate binding and product release
www.nature.com/articles/s41...
Our latest article is now published online! In collaboration with @thompson-lab.bsky.social, Marc Garcia-Borràs, and @ferranfeixas.bsky.social.
Distal Mutations in a Designed Retro-Aldolase Alter Loop Dynamics to Shift and Accelerate the Rate-Limiting Step
pubs.acs.org/doi/full/10....
Distal Mutations in a Designed Retro-Aldolase Alter Loop Dynamics to Shift and Accelerate the Rate-Limiting Step
pubs.acs.org/doi/full/10....

Distal Mutations in a Designed Retro-Aldolase Alter Loop Dynamics to Shift and Accelerate the Rate-Limiting Step
Amino acid residues distant from an enzyme’s active site are known to influence catalysis, but their mechanistic contributions to the catalytic cycle remain poorly understood. Here, we investigate the...
pubs.acs.org
August 26, 2025 at 12:41 PM
Our latest article is now published online! In collaboration with @thompson-lab.bsky.social, Marc Garcia-Borràs, and @ferranfeixas.bsky.social.
Distal Mutations in a Designed Retro-Aldolase Alter Loop Dynamics to Shift and Accelerate the Rate-Limiting Step
pubs.acs.org/doi/full/10....
Distal Mutations in a Designed Retro-Aldolase Alter Loop Dynamics to Shift and Accelerate the Rate-Limiting Step
pubs.acs.org/doi/full/10....
In collaboration with @birtehoecker.bsky.social, we’ve unlocked enzymatic activity in a minimal de novo TIM barrel by designing a custom lid for catalysis! 🧵👇
#ProteinDesign #EnzymeDesign
Customizing the Structure of a Minimal TIM Barrel to Create a De Novo Enzyme
www.biorxiv.org/content/10.1...
#ProteinDesign #EnzymeDesign
Customizing the Structure of a Minimal TIM Barrel to Create a De Novo Enzyme
www.biorxiv.org/content/10.1...

Customizing the Structure of a Minimal TIM Barrel to Craft a De Novo Enzyme
The TIM barrel is the most prevalent fold in natural enzymes, supporting efficient catalysis of diverse chemical reactions. While de novo TIM barrels have been successfully designed, their minimalisti...
www.biorxiv.org
July 29, 2025 at 6:33 PM
In collaboration with @birtehoecker.bsky.social, we’ve unlocked enzymatic activity in a minimal de novo TIM barrel by designing a custom lid for catalysis! 🧵👇
#ProteinDesign #EnzymeDesign
Customizing the Structure of a Minimal TIM Barrel to Create a De Novo Enzyme
www.biorxiv.org/content/10.1...
#ProteinDesign #EnzymeDesign
Customizing the Structure of a Minimal TIM Barrel to Create a De Novo Enzyme
www.biorxiv.org/content/10.1...
Reposted by Roberto Chica Lab
Join us! We are looking for a new team member (PhD student) with strong background in organic chemistry.
🙏 RETWEET (We want to recruit internationally)
Organic chemistry meets #DirectedEvolution
Highly interdisciplinary & passionate research group
uni-bielefeld.hr4you.org/job/view/433...
🙏 RETWEET (We want to recruit internationally)
Organic chemistry meets #DirectedEvolution
Highly interdisciplinary & passionate research group
uni-bielefeld.hr4you.org/job/view/433...

Research Position (PhD) in organic chemistry and b...
<div style="text-align: justify;">The research group „Organic Chemistry and&nb...
uni-bielefeld.hr4you.org
June 27, 2025 at 9:13 PM
Join us! We are looking for a new team member (PhD student) with strong background in organic chemistry.
🙏 RETWEET (We want to recruit internationally)
Organic chemistry meets #DirectedEvolution
Highly interdisciplinary & passionate research group
uni-bielefeld.hr4you.org/job/view/433...
🙏 RETWEET (We want to recruit internationally)
Organic chemistry meets #DirectedEvolution
Highly interdisciplinary & passionate research group
uni-bielefeld.hr4you.org/job/view/433...
Reposted by Roberto Chica Lab
Protein Engineering, Design & Selection (PEDS) invites contributions to a Special Collection titled, “Non-Canonical Amino Acids", with guest editors Prof. Huiwang Ai (Virginia) and Prof. Peng Chen (Peking). Send us your best work!
academic.oup.com/peds/pages/c...
academic.oup.com/peds/pages/c...
May 15, 2025 at 12:25 AM
Protein Engineering, Design & Selection (PEDS) invites contributions to a Special Collection titled, “Non-Canonical Amino Acids", with guest editors Prof. Huiwang Ai (Virginia) and Prof. Peng Chen (Peking). Send us your best work!
academic.oup.com/peds/pages/c...
academic.oup.com/peds/pages/c...
Reposted by Roberto Chica Lab
Super excited to share a new preprint from our lab on design of small-molecule binding proteins using neural networks! The paper has a bit of everything. A new graph neural network, new design algorithms, and experimental validation. www.biorxiv.org/content/10.1...
🧵🧪
🧵🧪

Zero-shot design of drug-binding proteins via neural selection-expansion
Computational design of molecular recognition remains challenging despite advances in deep learning. The design of proteins that bind to small molecules has been particularly difficult because it requ...
www.biorxiv.org
April 28, 2025 at 3:22 PM
Super excited to share a new preprint from our lab on design of small-molecule binding proteins using neural networks! The paper has a bit of everything. A new graph neural network, new design algorithms, and experimental validation. www.biorxiv.org/content/10.1...
🧵🧪
🧵🧪
Reposted by Roberto Chica Lab
Guess what? TPS has extended the deadline to March 19 to submit abstracts for poster presentations and speaking opportunities at our 39th Annual Symposium. Join us in San Francisco June 26 - 29 for 3.5 days of scientific talks.
hashtag#proteinscience hashtag#annualsymposium
lnkd.in/g7VKqX7C
hashtag#proteinscience hashtag#annualsymposium
lnkd.in/g7VKqX7C

March 6, 2025 at 6:27 PM
Guess what? TPS has extended the deadline to March 19 to submit abstracts for poster presentations and speaking opportunities at our 39th Annual Symposium. Join us in San Francisco June 26 - 29 for 3.5 days of scientific talks.
hashtag#proteinscience hashtag#annualsymposium
lnkd.in/g7VKqX7C
hashtag#proteinscience hashtag#annualsymposium
lnkd.in/g7VKqX7C
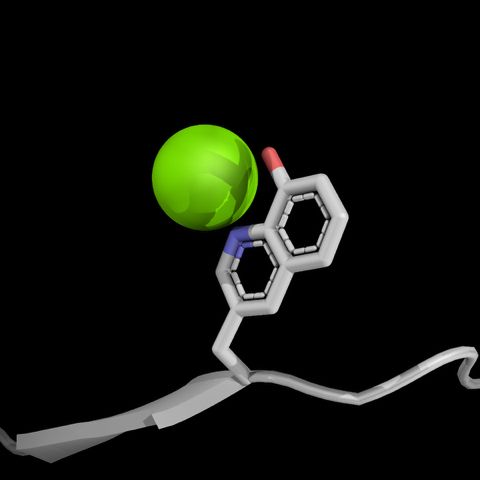
How do mutations far from an enzyme's active site influence catalysis? 🤔
Part 2: In collaboration with @fraserlab.bsky.social and @silviaosuna.bsky.social, we investigated this question using de novo Kemp eliminases, revealing effects of distal mutations on the catalytic cycle. 🧵 (1/6)
Part 2: In collaboration with @fraserlab.bsky.social and @silviaosuna.bsky.social, we investigated this question using de novo Kemp eliminases, revealing effects of distal mutations on the catalytic cycle. 🧵 (1/6)
February 28, 2025 at 5:09 PM
How do mutations far from an enzyme's active site influence catalysis? 🤔
Part 2: In collaboration with @fraserlab.bsky.social and @silviaosuna.bsky.social, we investigated this question using de novo Kemp eliminases, revealing effects of distal mutations on the catalytic cycle. 🧵 (1/6)
Part 2: In collaboration with @fraserlab.bsky.social and @silviaosuna.bsky.social, we investigated this question using de novo Kemp eliminases, revealing effects of distal mutations on the catalytic cycle. 🧵 (1/6)
Reposted by Roberto Chica Lab
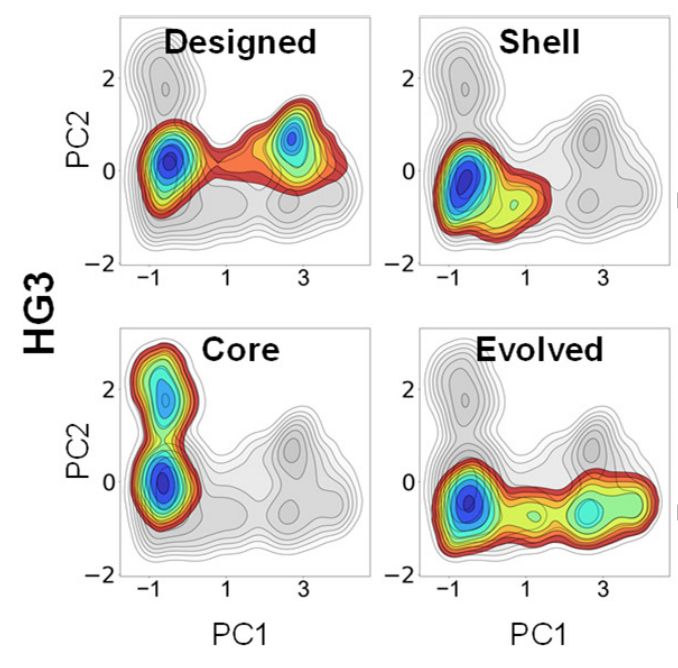
Whereas beneficial active site mutations to enzymes often improve the chemical transformation itself by preorganizing the active site, mutations to second-shell residues instead tune steps like product release by modifying the broader conformational ensemble www.biorxiv.org/content/10.1...
February 28, 2025 at 9:35 AM
Whereas beneficial active site mutations to enzymes often improve the chemical transformation itself by preorganizing the active site, mutations to second-shell residues instead tune steps like product release by modifying the broader conformational ensemble www.biorxiv.org/content/10.1...
How do distal mutations contribute to enzyme catalysis?
Our new study, in collaboration with @thompson-lab.bsky.social, Marc Garcia-Borràs, and @ferranfeixas.bsky.social, reveals how they optimize structural dynamics & local electric fields to drive the catalytic cycle. 🧵👇
Our new study, in collaboration with @thompson-lab.bsky.social, Marc Garcia-Borràs, and @ferranfeixas.bsky.social, reveals how they optimize structural dynamics & local electric fields to drive the catalytic cycle. 🧵👇
February 27, 2025 at 9:16 PM
How do distal mutations contribute to enzyme catalysis?
Our new study, in collaboration with @thompson-lab.bsky.social, Marc Garcia-Borràs, and @ferranfeixas.bsky.social, reveals how they optimize structural dynamics & local electric fields to drive the catalytic cycle. 🧵👇
Our new study, in collaboration with @thompson-lab.bsky.social, Marc Garcia-Borràs, and @ferranfeixas.bsky.social, reveals how they optimize structural dynamics & local electric fields to drive the catalytic cycle. 🧵👇
Reposted by Roberto Chica Lab
Join us this summer for 3.5 days for science! The Protein Society's 39th Annual Symposium takes place June 26 - 29, in beautiful San Francisco. #proteinscience #annualsymposium #proteinsociety
mailchi.mp/proteinsocie...
mailchi.mp/proteinsocie...
Register Today for PS39
mailchi.mp
February 5, 2025 at 2:14 PM
Join us this summer for 3.5 days for science! The Protein Society's 39th Annual Symposium takes place June 26 - 29, in beautiful San Francisco. #proteinscience #annualsymposium #proteinsociety
mailchi.mp/proteinsocie...
mailchi.mp/proteinsocie...
Reposted by Roberto Chica Lab
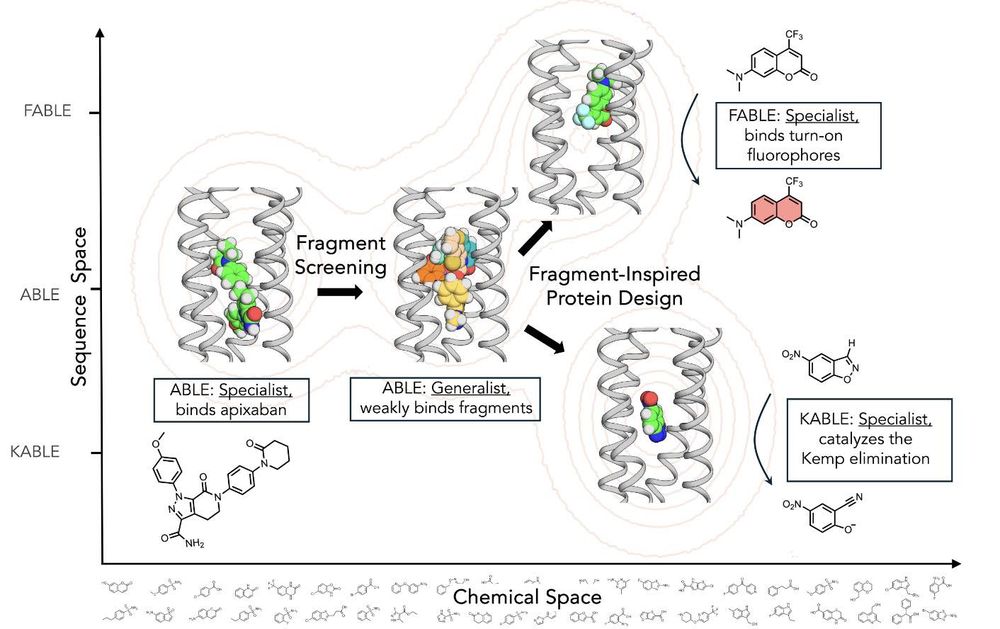
Thrilled to take a break from doom and gloom to share our latest work collaborating with the DeGrado lab! We took a de novo designed protein, screened it for ligand binding using X-rays, and used the hits to evolve two wildly different functions: fluorescent turn-on and Kemp elimination catalysis. 🧵
January 31, 2025 at 6:20 PM
Thrilled to take a break from doom and gloom to share our latest work collaborating with the DeGrado lab! We took a de novo designed protein, screened it for ligand binding using X-rays, and used the hits to evolve two wildly different functions: fluorescent turn-on and Kemp elimination catalysis. 🧵
Reposted by Roberto Chica Lab
Customizing the Structure of a Minimal TIM Barrel to Craft a De Novo Enzyme https://www.biorxiv.org/content/10.1101/2025.01.28.635154v1
January 30, 2025 at 2:46 AM
Customizing the Structure of a Minimal TIM Barrel to Craft a De Novo Enzyme https://www.biorxiv.org/content/10.1101/2025.01.28.635154v1
Reposted by Roberto Chica Lab
Distal mutations in a designed retro-aldolase alter loop dynamics to shift and accelerate the rate-limiting step https://www.biorxiv.org/content/10.1101/2025.01.26.634918v1
January 27, 2025 at 3:46 AM
Distal mutations in a designed retro-aldolase alter loop dynamics to shift and accelerate the rate-limiting step https://www.biorxiv.org/content/10.1101/2025.01.26.634918v1
Reposted by Roberto Chica Lab
Bluetorial-A dream and a bit of a nightmare
Serving as Editor-in-Chief at Science was fascinating. I greatly enjoyed working with talented and committed editorial, news, graphics, and production staff. But the inside look into scientific publishing and AAAS was also deeply disillusioning.
Serving as Editor-in-Chief at Science was fascinating. I greatly enjoyed working with talented and committed editorial, news, graphics, and production staff. But the inside look into scientific publishing and AAAS was also deeply disillusioning.

a cartoon says hey everybody an old man 's talking while bart simpson looks on
ALT: a cartoon says hey everybody an old man 's talking while bart simpson looks on
media.tenor.com
January 21, 2025 at 5:56 PM
Bluetorial-A dream and a bit of a nightmare
Serving as Editor-in-Chief at Science was fascinating. I greatly enjoyed working with talented and committed editorial, news, graphics, and production staff. But the inside look into scientific publishing and AAAS was also deeply disillusioning.
Serving as Editor-in-Chief at Science was fascinating. I greatly enjoyed working with talented and committed editorial, news, graphics, and production staff. But the inside look into scientific publishing and AAAS was also deeply disillusioning.
Looking forward to The Protein Society's 39th Annual Symposium!
Honored to co-chair the program planning committee with Margie Sunde, alongside committee stars @clvizcarra.bsky.social and Gary Zhang.
Explore the program here:
www.proteinsociety.org/program
Honored to co-chair the program planning committee with Margie Sunde, alongside committee stars @clvizcarra.bsky.social and Gary Zhang.
Explore the program here:
www.proteinsociety.org/program
January 17, 2025 at 8:51 PM
Looking forward to The Protein Society's 39th Annual Symposium!
Honored to co-chair the program planning committee with Margie Sunde, alongside committee stars @clvizcarra.bsky.social and Gary Zhang.
Explore the program here:
www.proteinsociety.org/program
Honored to co-chair the program planning committee with Margie Sunde, alongside committee stars @clvizcarra.bsky.social and Gary Zhang.
Explore the program here:
www.proteinsociety.org/program
Reposted by Roberto Chica Lab
Mireia Solà Colom gave an incredible seminar at @bpdmc.bsky.social last week, and I'm very excited for her to share some details about what we've built at @aiproteins.bsky.social and how well our #miniproteins perform as therapeutics
for the folks who weren't able to join for the brilliant presentation by Mireia Solà Colom last week to cap off our 2024 seminar series, you can check out the recording 👇
youtu.be/e6oZgGWjD_0
youtu.be/e6oZgGWjD_0
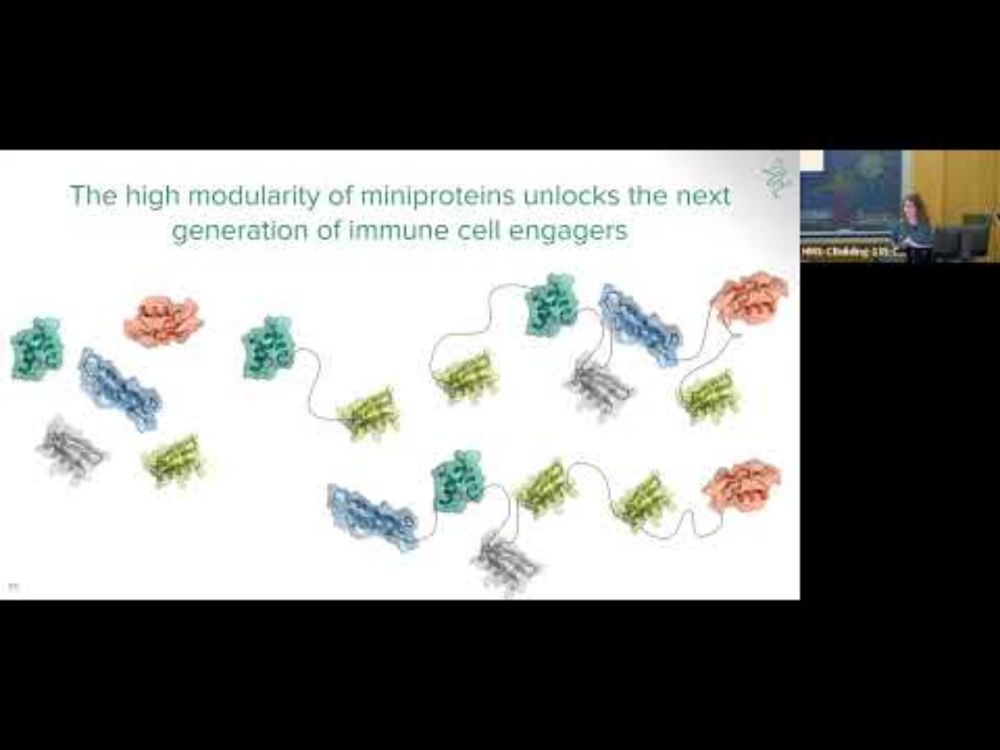
De novo design of miniprotein-based natural killer cell engagers
YouTube video by Boston Protein Design and Modeling Club
youtu.be
December 17, 2024 at 9:21 PM
Mireia Solà Colom gave an incredible seminar at @bpdmc.bsky.social last week, and I'm very excited for her to share some details about what we've built at @aiproteins.bsky.social and how well our #miniproteins perform as therapeutics
Reposted by Roberto Chica Lab
There is no "committee on science" that will impose a new order or set of incentives. No one is going to "fix" peer review or incentivize it differently in one fell swoop.
We have to BE the change - the tools already exist for everyone at every level to make an impact and change the incentives
We have to BE the change - the tools already exist for everyone at every level to make an impact and change the incentives
💡💥Here’s a modest proposal I’ve been pondering for a while on how to change the PEER REVIEW process and start more community-driven SCIENCE FUNDING. Lots of feedback/discussion welcomed! Other variants of the idea might exist already—not claiming originality. (a long 🧵) (1/9)
December 15, 2024 at 4:35 AM
There is no "committee on science" that will impose a new order or set of incentives. No one is going to "fix" peer review or incentivize it differently in one fell swoop.
We have to BE the change - the tools already exist for everyone at every level to make an impact and change the incentives
We have to BE the change - the tools already exist for everyone at every level to make an impact and change the incentives
Reposted by Roberto Chica Lab
📣 Save the dates 📅
We are organizing a Benzon Symposium on "Protein structure prediction and design" with what I think is an amazing set of speakers
Meeting will take place in Copenhagen 🇩🇰 on Sept. 1–4, 2025, and abstract submission will open in March (benzon-foundation.dk/benzon-sympo...)
We are organizing a Benzon Symposium on "Protein structure prediction and design" with what I think is an amazing set of speakers
Meeting will take place in Copenhagen 🇩🇰 on Sept. 1–4, 2025, and abstract submission will open in March (benzon-foundation.dk/benzon-sympo...)

December 13, 2024 at 2:50 PM
📣 Save the dates 📅
We are organizing a Benzon Symposium on "Protein structure prediction and design" with what I think is an amazing set of speakers
Meeting will take place in Copenhagen 🇩🇰 on Sept. 1–4, 2025, and abstract submission will open in March (benzon-foundation.dk/benzon-sympo...)
We are organizing a Benzon Symposium on "Protein structure prediction and design" with what I think is an amazing set of speakers
Meeting will take place in Copenhagen 🇩🇰 on Sept. 1–4, 2025, and abstract submission will open in March (benzon-foundation.dk/benzon-sympo...)
Reposted by Roberto Chica Lab
Many industrially-important reactions (e.g. lipase-catalyzed biofuel production) involve substrates poorly soluble in water. How can we screen enzymes for these reactions? Samuel Thompson invented a new 'triple emulsion' platform for biphasic screening (1/n):
onlinelibrary.wiley.com/doi/full/10....
onlinelibrary.wiley.com/doi/full/10....

FACS‐Sortable Triple Emulsion Picoreactors for Screening Reactions in Biphasic Environments
Biphasic reactions can enhance reactions where the catalyst and substrate have different solvent preferences. This study optimizes triple emulsion picoreactors that encapsulate a biphasic solvent env...
onlinelibrary.wiley.com
December 13, 2024 at 1:54 AM
Many industrially-important reactions (e.g. lipase-catalyzed biofuel production) involve substrates poorly soluble in water. How can we screen enzymes for these reactions? Samuel Thompson invented a new 'triple emulsion' platform for biphasic screening (1/n):
onlinelibrary.wiley.com/doi/full/10....
onlinelibrary.wiley.com/doi/full/10....
Reposted by Roberto Chica Lab
Please share this PhD opportunity in my new lab in Bristol, using protein design/ engineering to understand and build molecular motors to control intracellular transport. Working with @markdodding.bsky.social and Prof Dek Woolfson.
www.findaphd.com/phds/project...
www.swbio.ac.uk/biomolecular...
www.findaphd.com/phds/project...
www.swbio.ac.uk/biomolecular...
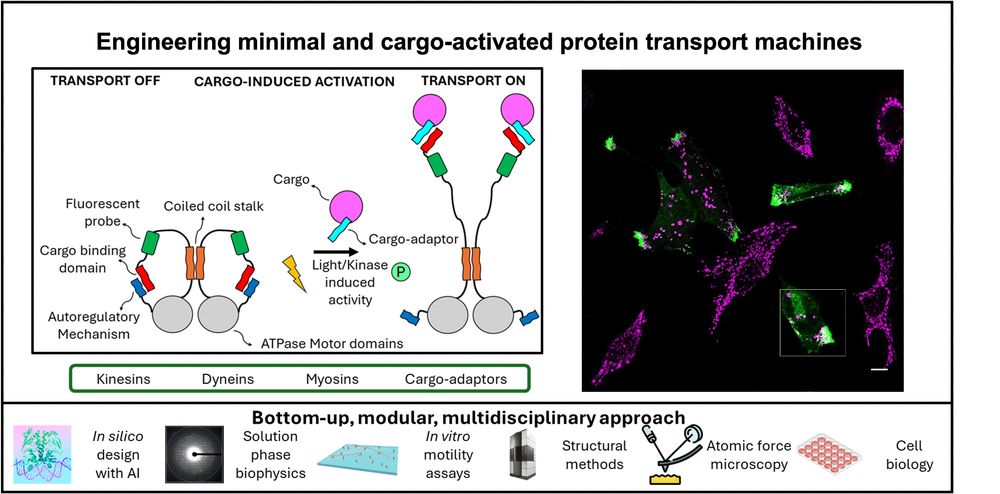
November 28, 2024 at 4:55 PM
Please share this PhD opportunity in my new lab in Bristol, using protein design/ engineering to understand and build molecular motors to control intracellular transport. Working with @markdodding.bsky.social and Prof Dek Woolfson.
www.findaphd.com/phds/project...
www.swbio.ac.uk/biomolecular...
www.findaphd.com/phds/project...
www.swbio.ac.uk/biomolecular...
Reposted by Roberto Chica Lab
🚨Announcing NetSci: a super fast tool to compute correlated motion in biomolecules
pubs.acs.org/doi/10.1021/...
@pabloarantes.bsky.social & team also made it into a Colab notebook:
colab.research.google.com/drive/1GGJKr...
Give it a try & send feedback!
pubs.acs.org/doi/10.1021/...
@pabloarantes.bsky.social & team also made it into a Colab notebook:
colab.research.google.com/drive/1GGJKr...
Give it a try & send feedback!

NetSci: A Library for High Performance Biomolecular Simulation Network Analysis Computation
We present the NetSci program–an open-source scientific software package designed for estimating mutual information (MI) between data sets using GPU acceleration and a k-nearest-neighbor algorithm. This approach significantly enhances calculation speed, achieving improvements of several orders of magnitude over traditional CPU-based methods, with data set size limits dictated only by available hardware. To validate NetSci, we accurately compute MI for an analytically verifiable two-dimensional Gaussian distribution and replicate the generalized correlation (GC) analysis previously conducted on the B1 domain of protein G. We also apply NetSci to molecular dynamics simulations of the Sarcoendoplasmic Reticulum Calcium-ATPase (SERCA) pump, exploring the allosteric mechanisms and pathways influenced by ATP and 2′-deoxy-ATP (dATP) binding. Our analysis reveals distinct allosteric effects induced by ATP compared to dATP, with predicted information pathways from the bound nucleotide to the calcium-binding domain differing based on the nucleotide involved. NetSci proves to be a valuable tool for estimating MI and GC in various data sets and is particularly effective for analyzing intraprotein communication and information transfer.
pubs.acs.org
November 18, 2024 at 6:49 PM
🚨Announcing NetSci: a super fast tool to compute correlated motion in biomolecules
pubs.acs.org/doi/10.1021/...
@pabloarantes.bsky.social & team also made it into a Colab notebook:
colab.research.google.com/drive/1GGJKr...
Give it a try & send feedback!
pubs.acs.org/doi/10.1021/...
@pabloarantes.bsky.social & team also made it into a Colab notebook:
colab.research.google.com/drive/1GGJKr...
Give it a try & send feedback!
Reposted by Roberto Chica Lab
Happy to announce our latest article on #enzymedesign, in collaboration with Mike Thompson's group! Check it out in JACS:
Design of Efficient Artificial Enzymes Using Crystallographically Enhanced Conformational Sampling
pubs.acs.org/doi/full/10....
Design of Efficient Artificial Enzymes Using Crystallographically Enhanced Conformational Sampling
pubs.acs.org/doi/full/10....
March 28, 2024 at 2:49 PM
Happy to announce our latest article on #enzymedesign, in collaboration with Mike Thompson's group! Check it out in JACS:
Design of Efficient Artificial Enzymes Using Crystallographically Enhanced Conformational Sampling
pubs.acs.org/doi/full/10....
Design of Efficient Artificial Enzymes Using Crystallographically Enhanced Conformational Sampling
pubs.acs.org/doi/full/10....
Reposted by Roberto Chica Lab
go.bsky.app/BoXpqQp
Computational protein design starter pack. Let me know if I missed you!
Computational protein design starter pack. Let me know if I missed you!
November 17, 2024 at 11:55 AM
go.bsky.app/BoXpqQp
Computational protein design starter pack. Let me know if I missed you!
Computational protein design starter pack. Let me know if I missed you!
Happy to announce our latest article on #enzymedesign, in collaboration with Mike Thompson's group! Check it out in JACS:
Design of Efficient Artificial Enzymes Using Crystallographically Enhanced Conformational Sampling
pubs.acs.org/doi/full/10....
Design of Efficient Artificial Enzymes Using Crystallographically Enhanced Conformational Sampling
pubs.acs.org/doi/full/10....
March 28, 2024 at 2:49 PM
Happy to announce our latest article on #enzymedesign, in collaboration with Mike Thompson's group! Check it out in JACS:
Design of Efficient Artificial Enzymes Using Crystallographically Enhanced Conformational Sampling
pubs.acs.org/doi/full/10....
Design of Efficient Artificial Enzymes Using Crystallographically Enhanced Conformational Sampling
pubs.acs.org/doi/full/10....

