
ARC DECRA Fellow at Monash University 🇦🇺.
Structural biologist. Author of WIGGLE. He/Him. 🏳️🌈

This recognition is a huge testament to the amazing work of our entire team. 🙌💥🎉
Photography and video: @vicgovau.bsky.social
www.youtube.com/watch?v=9_Ht...
www.biorxiv.org/content/10.1...
Boltz-2 is an excellent open source alternative to AlphaFold3. However, high VRAM use restricts modeling large complexes. Using careful memory management, we increase the Boltz-2 size limit by >60% while maintaining execution speed.

www.biorxiv.org/content/10.1...
Boltz-2 is an excellent open source alternative to AlphaFold3. However, high VRAM use restricts modeling large complexes. Using careful memory management, we increase the Boltz-2 size limit by >60% while maintaining execution speed.

🌐 evedesign.bio
🌐 evedesign.bio
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Today, we share our preprint “Generative design of novel bacteriophages with genome language models”, where we validate the first, functional AI-generated genomes 🧵
Today, we share our preprint “Generative design of novel bacteriophages with genome language models”, where we validate the first, functional AI-generated genomes 🧵
It combines sensitive Foldseek structure search and conserved neighborhood detection to discover functionally-associated gene clusters in prokaryotic & viral genomes.
1/6🧵
@ruoshiz.bsky.social @milot.bsky.social
www.nature.com/articles/s41...

It combines sensitive Foldseek structure search and conserved neighborhood detection to discover functionally-associated gene clusters in prokaryotic & viral genomes.
1/6🧵

We introduce #APEX - an open-source, automated pipeline for recombinant protein production in E. coli, developed by our super talented Martyna Kasprzyk!
Check it out: pubs.acs.org/doi/10.1021/...
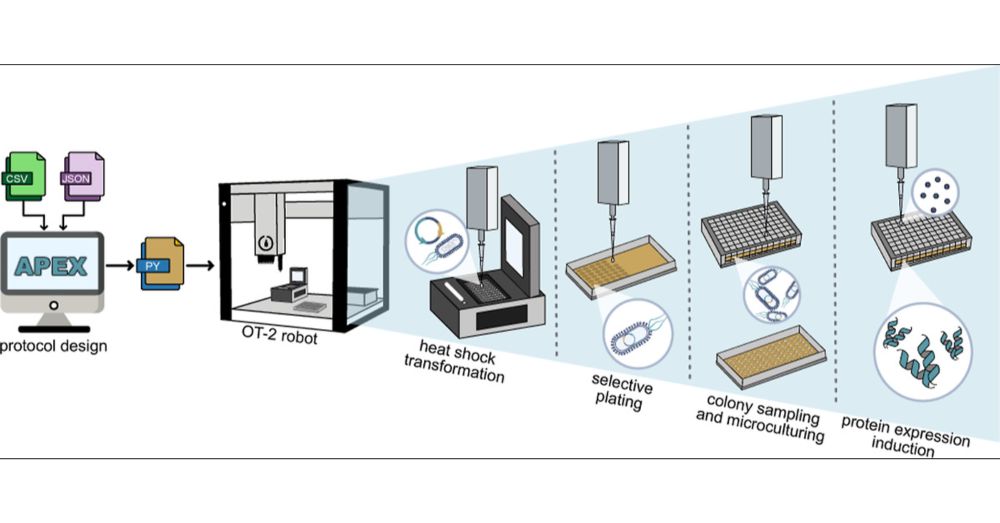
We introduce #APEX - an open-source, automated pipeline for recombinant protein production in E. coli, developed by our super talented Martyna Kasprzyk!
Check it out: pubs.acs.org/doi/10.1021/...
Congrats Hannah Ochner and authors on this important paper! Strong collaboration with @kiranrpatil.bsky.social
www.biorxiv.org/cgi/content/...
@mrclmb.bsky.social @wellcometrust.bsky.social

Congrats Hannah Ochner and authors on this important paper! Strong collaboration with @kiranrpatil.bsky.social
www.biorxiv.org/cgi/content/...
@mrclmb.bsky.social @wellcometrust.bsky.social
This recognition is a huge testament to the amazing work of our entire team. 🙌💥🎉
Photography and video: @vicgovau.bsky.social
www.youtube.com/watch?v=9_Ht...

This recognition is a huge testament to the amazing work of our entire team. 🙌💥🎉
Photography and video: @vicgovau.bsky.social
www.youtube.com/watch?v=9_Ht...
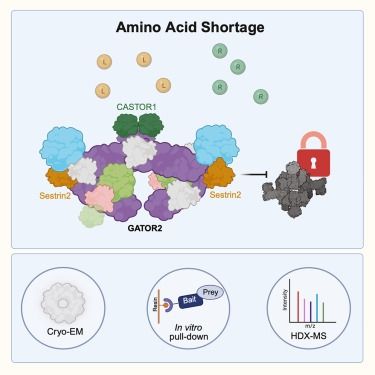
biorxiv.org/content/10.110
biorxiv.org/content/10.110
@wehi-research.bsky.social
@wehi-research.bsky.social
Super grateful to have played a small role in this project - congrats to lead/corr authors Oliver, Benjamin, and Markus!
www.nature.com/articles/s41...
#alphafold #evolution #genomics

www.nature.com/articles/s41...
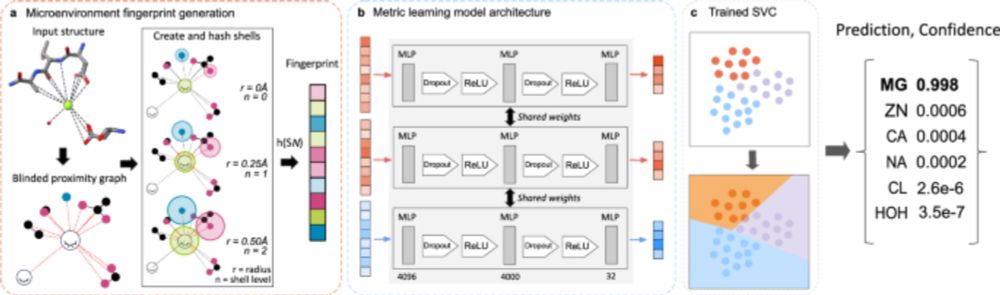
www.nature.com/articles/s41...

