Alex Arzamasov
@arzamasovalex.bsky.social
Postdoc in Osterman lab at SBP Medical Discovery Institute.
Interested in the functional annotation of genomic and metagenomic data, carbohydrate metabolism, #bifidobacteria
Interested in the functional annotation of genomic and metagenomic data, carbohydrate metabolism, #bifidobacteria
Pinned
Alex Arzamasov
@arzamasovalex.bsky.social
· Sep 30
It’s been a bit over 2 months since the main results of my PhD work on reconstructing carbohydrate utilization pathways in human bifidobacteria were finally published; so I guess it’s a good time to update the previous thread (1/7) www.nature.com/articles/s41...
Some cool data on bifidobacteria and their relatives in honeybees!
Happy to announce that our comparative metagenomic analysis of the gut microbiota of five honeybee species - spearheaded by @aiswarya.bsky.social - is finally published in a peer-reviewed journal! rdcu.be/eKMCs @fbm-unil.bsky.social @dmf-unil.bsky.social 🧵👇

Evolution of gut microbiota across honeybee species revealed by comparative metagenomics
Nature Communications - Here, the authors perform metagenomic analysis of honeybee gut microbes, uncovering many previously unidentified species and host specific differences in composition and...
rdcu.be
October 14, 2025 at 9:48 PM
Some cool data on bifidobacteria and their relatives in honeybees!
Reposted by Alex Arzamasov
Happy to announce that our comparative metagenomic analysis of the gut microbiota of five honeybee species - spearheaded by @aiswarya.bsky.social - is finally published in a peer-reviewed journal! rdcu.be/eKMCs @fbm-unil.bsky.social @dmf-unil.bsky.social 🧵👇

Evolution of gut microbiota across honeybee species revealed by comparative metagenomics
Nature Communications - Here, the authors perform metagenomic analysis of honeybee gut microbes, uncovering many previously unidentified species and host specific differences in composition and...
rdcu.be
October 14, 2025 at 7:04 PM
Happy to announce that our comparative metagenomic analysis of the gut microbiota of five honeybee species - spearheaded by @aiswarya.bsky.social - is finally published in a peer-reviewed journal! rdcu.be/eKMCs @fbm-unil.bsky.social @dmf-unil.bsky.social 🧵👇
It’s been a bit over 2 months since the main results of my PhD work on reconstructing carbohydrate utilization pathways in human bifidobacteria were finally published; so I guess it’s a good time to update the previous thread (1/7) www.nature.com/articles/s41...
September 30, 2025 at 9:01 AM
It’s been a bit over 2 months since the main results of my PhD work on reconstructing carbohydrate utilization pathways in human bifidobacteria were finally published; so I guess it’s a good time to update the previous thread (1/7) www.nature.com/articles/s41...
Reposted by Alex Arzamasov
Excited to share our @cp-cellhostmicrobe.bsky.social led by Magda showing how Bif has co-evolved with different animal hosts 🐒🐭🐷🐦
Key takeaways:
🔹 Host ancestry + diet shape Bif evolution
🔹 Mammals enriched for carb-busting enzymes
🔹 Untapped diversity in non-human hosts = new probiotic potential
Key takeaways:
🔹 Host ancestry + diet shape Bif evolution
🔹 Mammals enriched for carb-busting enzymes
🔹 Untapped diversity in non-human hosts = new probiotic potential
Bifido adaptations across hosts
@halllab.bsky.social survey insects, reptiles, birds & mammals to uncover how Bifidobacterium adapts to hosts. Bifidobacterium & host exhibit strong co-phylogenetic associations, driven by vertical transmission & dietary selection
www.cell.com/cell-host-mi...
@halllab.bsky.social survey insects, reptiles, birds & mammals to uncover how Bifidobacterium adapts to hosts. Bifidobacterium & host exhibit strong co-phylogenetic associations, driven by vertical transmission & dietary selection
www.cell.com/cell-host-mi...

Host-specific microbiome and genomic signatures in Bifidobacterium reveal co-evolutionary and functional adaptations across diverse animal hosts
Bifidobacterium are beneficial members of the microbiota across animal hosts. Kujawska
et al. demonstrate that bifidobacteria have likely co-evolved more closely with mammals,
particularly primates, a...
www.cell.com
September 11, 2025 at 2:49 PM
Excited to share our @cp-cellhostmicrobe.bsky.social led by Magda showing how Bif has co-evolved with different animal hosts 🐒🐭🐷🐦
Key takeaways:
🔹 Host ancestry + diet shape Bif evolution
🔹 Mammals enriched for carb-busting enzymes
🔹 Untapped diversity in non-human hosts = new probiotic potential
Key takeaways:
🔹 Host ancestry + diet shape Bif evolution
🔹 Mammals enriched for carb-busting enzymes
🔹 Untapped diversity in non-human hosts = new probiotic potential
Reposted by Alex Arzamasov
Our study developing a skin metatranscriptomics protocol is now out in @natbiotech.nature.com!
We finally have the ability to study microbial activity on skin and identify key functional genes playing a role in diseases.
Amazing team of Chia Minghao and Amanda Ng 👏
nature.com/articles/s41...
We finally have the ability to study microbial activity on skin and identify key functional genes playing a role in diseases.
Amazing team of Chia Minghao and Amanda Ng 👏
nature.com/articles/s41...
August 30, 2025 at 2:17 AM
Our study developing a skin metatranscriptomics protocol is now out in @natbiotech.nature.com!
We finally have the ability to study microbial activity on skin and identify key functional genes playing a role in diseases.
Amazing team of Chia Minghao and Amanda Ng 👏
nature.com/articles/s41...
We finally have the ability to study microbial activity on skin and identify key functional genes playing a role in diseases.
Amazing team of Chia Minghao and Amanda Ng 👏
nature.com/articles/s41...
Very nice paper that deciphers the transport mechanisms of fucosylated lacto-N-biose in Bifidobacterium longum subsp. infantis
journals.asm.org/doi/full/10....
journals.asm.org/doi/full/10....

Uptake of fucosylated type I human milk oligosaccharide blocks by Bifidobacterium longum subsp. infantis | mBio
The assembly of the gut microbiota in early life is critical to the health trajectory
of human hosts. Breast feeding selects for a Bifidobacterium-rich community, adapted to efficiently utilize human ...
journals.asm.org
July 26, 2025 at 4:19 AM
Very nice paper that deciphers the transport mechanisms of fucosylated lacto-N-biose in Bifidobacterium longum subsp. infantis
journals.asm.org/doi/full/10....
journals.asm.org/doi/full/10....
Reposted by Alex Arzamasov
High-throughput single-cell isolation of Bifidobacterium strains from the gut microbiome https://www.biorxiv.org/content/10.1101/2025.07.23.666462v1
July 25, 2025 at 4:17 AM
High-throughput single-cell isolation of Bifidobacterium strains from the gut microbiome https://www.biorxiv.org/content/10.1101/2025.07.23.666462v1
Reposted by Alex Arzamasov
New Pre-print! “PNGaseA-mediated N-glycan stripping from peptides by infant-derived Bifidobacterium bifidum”. This is the first manuscript from the Bifidobacterium and breast milk project🍼🤱👶in collaboration with van Sinderen and Lovering groups
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

PNGaseA-mediated N-glycan stripping from peptides by infant-derived Bifidobacterium bifidum
N-glycans are highly common sources of nutrition for human colonic-dwelling bacteria. These microbes have evolved a several methods to remove N-glycans from proteins; herein we describe the biochemica...
www.biorxiv.org
July 18, 2025 at 8:52 AM
New Pre-print! “PNGaseA-mediated N-glycan stripping from peptides by infant-derived Bifidobacterium bifidum”. This is the first manuscript from the Bifidobacterium and breast milk project🍼🤱👶in collaboration with van Sinderen and Lovering groups
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Alex Arzamasov
Great to see our manuscript on skDER & CiDDER - programs for selection of representative microbial genomes - now published.
Please give them a try and if you have any feature requests or issues, just let us know.
www.microbiologyresearch.org/content/jour...
github.com/raufs/skDER
Please give them a try and if you have any feature requests or issues, just let us know.
www.microbiologyresearch.org/content/jour...
github.com/raufs/skDER

skDER and CiDDER: two scalable approaches for microbial genome dereplication
An abundance of microbial genomes have been sequenced in the past two decades. For fundamental comparative genomic investigations, where the goal is to determine the major gain and loss events shaping...
www.microbiologyresearch.org
July 11, 2025 at 8:14 PM
Great to see our manuscript on skDER & CiDDER - programs for selection of representative microbial genomes - now published.
Please give them a try and if you have any feature requests or issues, just let us know.
www.microbiologyresearch.org/content/jour...
github.com/raufs/skDER
Please give them a try and if you have any feature requests or issues, just let us know.
www.microbiologyresearch.org/content/jour...
github.com/raufs/skDER
Reposted by Alex Arzamasov
The discovery of the missing Queuosine transporter is finally out. 15 years in the making with a combination of data mining and experimental validation performed by an international consortium funded by NIH and Irish agencies.
news.ufl.edu/2025/06/micr...
news.ufl.edu/2025/06/micr...

UF, Trinity College team cracks micronutrient mystery that could be key to brain health, cancer defense
Unlocking how our bodies absorb a micronutrient that we rely on for everything from healthy brain function to cancer defense.
news.ufl.edu
June 18, 2025 at 4:24 PM
The discovery of the missing Queuosine transporter is finally out. 15 years in the making with a combination of data mining and experimental validation performed by an international consortium funded by NIH and Irish agencies.
news.ufl.edu/2025/06/micr...
news.ufl.edu/2025/06/micr...
Reposted by Alex Arzamasov
A great blog post by @math-rachel.bsky.social on this preprint describing failures of an ML approach for gene annotation:
rachel.fast.ai/posts/2025-0...
www.biorxiv.org/content/10.1...
"how challenging (or even impossible) it can be to evaluate AI claims in work outside our own area of expertise"
rachel.fast.ai/posts/2025-0...
www.biorxiv.org/content/10.1...
"how challenging (or even impossible) it can be to evaluate AI claims in work outside our own area of expertise"

Rachel Thomas, PhD - Deep learning gets the glory, deep fact checking gets ignored
an AI researcher going back to school for immunology
rachel.fast.ai
June 4, 2025 at 4:29 PM
A great blog post by @math-rachel.bsky.social on this preprint describing failures of an ML approach for gene annotation:
rachel.fast.ai/posts/2025-0...
www.biorxiv.org/content/10.1...
"how challenging (or even impossible) it can be to evaluate AI claims in work outside our own area of expertise"
rachel.fast.ai/posts/2025-0...
www.biorxiv.org/content/10.1...
"how challenging (or even impossible) it can be to evaluate AI claims in work outside our own area of expertise"
I often flag genomic predictions of SCFA production that clash with “textbook” data. But sometimes it’s the experimental claims that raise eyebrows. This recent paper reports propionate (!) and butyrate (!!) production by Bif. longum ssp. infantis (it shouldn't make either
doi.org/10.3168/jds....
doi.org/10.3168/jds....

May 19, 2025 at 4:13 AM
I often flag genomic predictions of SCFA production that clash with “textbook” data. But sometimes it’s the experimental claims that raise eyebrows. This recent paper reports propionate (!) and butyrate (!!) production by Bif. longum ssp. infantis (it shouldn't make either
doi.org/10.3168/jds....
doi.org/10.3168/jds....
Great resource! A small note on Bifidobacterium hominis: the description mentions "predicted starch utilization and propionate production", but bifidobacteria don’t produce propionate, and this species lacks extracellular alpha-amylases for starch degradation. Hope this can be refined in the future!
Very happy that HiBC is out: rdcu.be/ekSCO 🔥 @natcomms.nature.com Excellent team spirit, and, as always, great driving factor by @tcahitch.bsky.social ⭐⭐⭐⭐ We hope that these bacterial isolates from the human gut will facilitate many studies by others... 😀
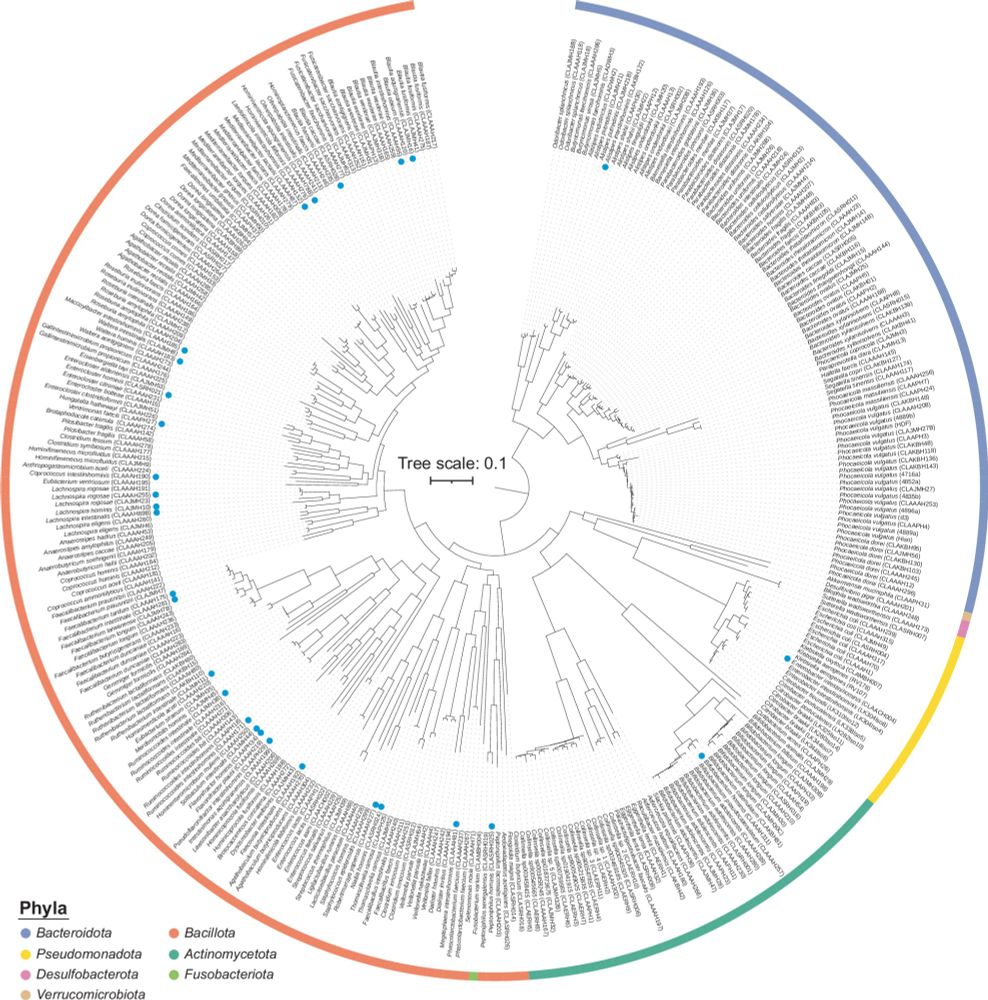
HiBC: a publicly available collection of bacterial strains isolated from the human gut
Nature Communications - Here, the authors present and characterise a collection of human gut bacteria including novel taxa associated with health conditions and a large diversity of plasmids. All...
rdcu.be
May 6, 2025 at 11:58 PM
Great resource! A small note on Bifidobacterium hominis: the description mentions "predicted starch utilization and propionate production", but bifidobacteria don’t produce propionate, and this species lacks extracellular alpha-amylases for starch degradation. Hope this can be refined in the future!
Reposted by Alex Arzamasov
Diversity and prevalence of Anaerostipes in the human gut microbiota https://www.biorxiv.org/content/10.1101/2025.05.01.651700v1
May 2, 2025 at 4:17 AM
Diversity and prevalence of Anaerostipes in the human gut microbiota https://www.biorxiv.org/content/10.1101/2025.05.01.651700v1
Reposted by Alex Arzamasov
I got hit by some rather sudden and extreme financial hardship so if anyone is in need of remote wetlab contract research, strictly BSL1, do let me know. Currently scrambling for gigs.
Plant, Bacterial, Archaeal Non-model Bioeng
Custom Lab Hardware
Turn Key Genetic Design
Please repost for reach 💚
Plant, Bacterial, Archaeal Non-model Bioeng
Custom Lab Hardware
Turn Key Genetic Design
Please repost for reach 💚
May 2, 2025 at 2:39 PM
I got hit by some rather sudden and extreme financial hardship so if anyone is in need of remote wetlab contract research, strictly BSL1, do let me know. Currently scrambling for gigs.
Plant, Bacterial, Archaeal Non-model Bioeng
Custom Lab Hardware
Turn Key Genetic Design
Please repost for reach 💚
Plant, Bacterial, Archaeal Non-model Bioeng
Custom Lab Hardware
Turn Key Genetic Design
Please repost for reach 💚
Reposted by Alex Arzamasov
Small amounts of misassembly can have disproportionate effects on pangenome-based metagenomic analyses journals.asm.org/doi/full/10.... #jcampubs
April 29, 2025 at 5:17 PM
Small amounts of misassembly can have disproportionate effects on pangenome-based metagenomic analyses journals.asm.org/doi/full/10.... #jcampubs
Reposted by Alex Arzamasov
I am very happy (and anxious) to share with you our most recent work in which we evaluated four of the most popular long-read assemblers,
www.biorxiv.org/content/10.1...
and tell you just a little bit about it in the following 🧵
www.biorxiv.org/content/10.1...
and tell you just a little bit about it in the following 🧵

Assemblies of long-read metagenomes suffer from diverse errors
Genomes from metagenomes have revolutionised our understanding of microbial diversity, ecology, and evolution, propelling advances in basic science, biomedicine, and biotechnology. Assembly algorithms...
www.biorxiv.org
April 28, 2025 at 8:08 AM
I am very happy (and anxious) to share with you our most recent work in which we evaluated four of the most popular long-read assemblers,
www.biorxiv.org/content/10.1...
and tell you just a little bit about it in the following 🧵
www.biorxiv.org/content/10.1...
and tell you just a little bit about it in the following 🧵
Reposted by Alex Arzamasov
GTDB release 10 based on RefSeq 226 (R10-RS226) is live at gtdb.ecogenomic.org. This release covers 732,475 genomes (22% increase) and has 143,6141 species clusters (37% increase). Release notes at: forum.gtdb.ecogenomic.org/t/announcing.... Release statistics at: gtdb.ecogenomic.org/stats/r226.
GTDB - Genome Taxonomy Database
The Genome Taxonomy Database (GTDB) is an initiative to establish a standardised microbial taxonomy based on genome phylogeny.
gtdb.ecogenomic.org
April 18, 2025 at 7:13 PM
GTDB release 10 based on RefSeq 226 (R10-RS226) is live at gtdb.ecogenomic.org. This release covers 732,475 genomes (22% increase) and has 143,6141 species clusters (37% increase). Release notes at: forum.gtdb.ecogenomic.org/t/announcing.... Release statistics at: gtdb.ecogenomic.org/stats/r226.
Nice paper led by Aruto Nakajima (Katayama lab) that provides mechanistic insights on 2'-fucosyllactose (2'FL) and other HMO utilization by Clostridium perfringens. There also some cool data on in vitro competition for 2'FL with different Bifidobacterium species
www.tandfonline.com/doi/full/10....
www.tandfonline.com/doi/full/10....

In vitro competition with Bifidobacterium strains impairs potentially pathogenic growth of Clostridium perfringens on 2′-fucosyllactose
Fortifying infant formula with human milk oligosaccharides, such as 2'-fucosyllactose (2'-FL), is a global trend. Previous studies have shown the inability of pathogenic gut microbes to utilize 2'-...
www.tandfonline.com
March 19, 2025 at 6:55 PM
Nice paper led by Aruto Nakajima (Katayama lab) that provides mechanistic insights on 2'-fucosyllactose (2'FL) and other HMO utilization by Clostridium perfringens. There also some cool data on in vitro competition for 2'FL with different Bifidobacterium species
www.tandfonline.com/doi/full/10....
www.tandfonline.com/doi/full/10....
Reposted by Alex Arzamasov
DNA-utilization loci enable exogenous DNA metabolism in gut Bacteroidales https://www.biorxiv.org/content/10.1101/2025.03.18.642099v1
March 19, 2025 at 4:23 AM
DNA-utilization loci enable exogenous DNA metabolism in gut Bacteroidales https://www.biorxiv.org/content/10.1101/2025.03.18.642099v1
Reposted by Alex Arzamasov
Interested in the influence of the infant microbiome on health? Quickly interrogating non-model organisms using omic approaches?
A new publication, describing genome-scale resources in bifidobacteria, is now available online: doi.org/10.1016/j.ce...
#microbiome #microsky
A new publication, describing genome-scale resources in bifidobacteria, is now available online: doi.org/10.1016/j.ce...
#microbiome #microsky

March 13, 2025 at 10:35 PM
Interested in the influence of the infant microbiome on health? Quickly interrogating non-model organisms using omic approaches?
A new publication, describing genome-scale resources in bifidobacteria, is now available online: doi.org/10.1016/j.ce...
#microbiome #microsky
A new publication, describing genome-scale resources in bifidobacteria, is now available online: doi.org/10.1016/j.ce...
#microbiome #microsky
New insights into the substrate-specificity of transporters in Bifidobacterium longum subsp. infantis
Uptake of fucosylated type I human milk oligosaccharide blocks by Bifidobacterium longum subsp. infantis https://www.biorxiv.org/content/10.1101/2025.02.07.637197v1
February 10, 2025 at 10:24 PM
New insights into the substrate-specificity of transporters in Bifidobacterium longum subsp. infantis
Very important work showing that many human gut bacteria can metabolize various human milk oligosaccharides (HMOs)
Super excited to share our latest research on how HMOs shape the post-weaned gut microbiota! Huge thanks to our incredible team, this wouldn’t have been possible without their dedication and expertise @emmaallen-vercoe.bsky.social @drlarsbode.bsky.social
Just out! Results of an incredible effort 👏 by @simonerenwick.bsky.social to demonstrate how different gut microbes and #gutmicrobiomes respond to human milk oligosaccharides (HMOs). Many more microbes respond to HMOs than you might think! Give it a read… links.springernature.com/f/a/DF0RtJaQ...
February 7, 2025 at 9:13 PM
Very important work showing that many human gut bacteria can metabolize various human milk oligosaccharides (HMOs)
Reposted by Alex Arzamasov
Super excited to share our latest research on how HMOs shape the post-weaned gut microbiota! Huge thanks to our incredible team, this wouldn’t have been possible without their dedication and expertise @emmaallen-vercoe.bsky.social @drlarsbode.bsky.social
Just out! Results of an incredible effort 👏 by @simonerenwick.bsky.social to demonstrate how different gut microbes and #gutmicrobiomes respond to human milk oligosaccharides (HMOs). Many more microbes respond to HMOs than you might think! Give it a read… links.springernature.com/f/a/DF0RtJaQ...
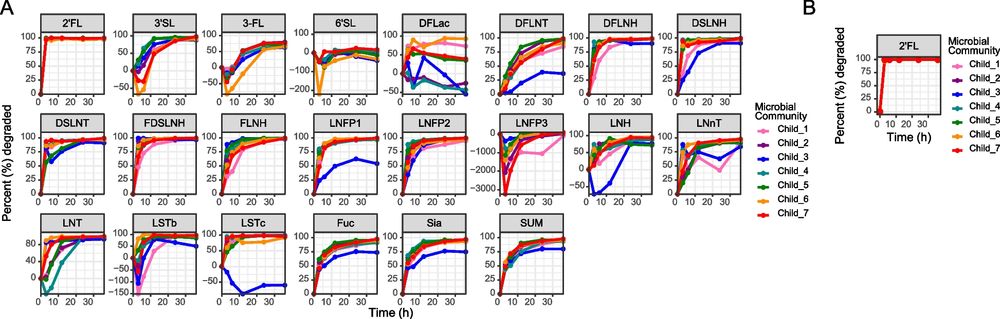
Modulating the developing gut microbiota with 2’-fucosyllactose and pooled human milk oligosaccharides - Microbiome
Background Synthetic human milk oligosaccharides (HMOs) are used to supplement infant formula despite limited understanding of their impact on the post-weaned developing gut microbiota. Here, we asses...
links.springernature.com
February 7, 2025 at 3:40 PM
Super excited to share our latest research on how HMOs shape the post-weaned gut microbiota! Huge thanks to our incredible team, this wouldn’t have been possible without their dedication and expertise @emmaallen-vercoe.bsky.social @drlarsbode.bsky.social
Reposted by Alex Arzamasov
Difficult to focus on science during these times, but if you have the bandwidth, our study on zol is now out in NAR: academic.oup.com/nar/article/...
GitHub: github.com/Kalan-Lab/zol
Please give the software a try and let us know if you have any feedback! And stay tuned for new features...
GitHub: github.com/Kalan-Lab/zol
Please give the software a try and let us know if you have any feedback! And stay tuned for new features...
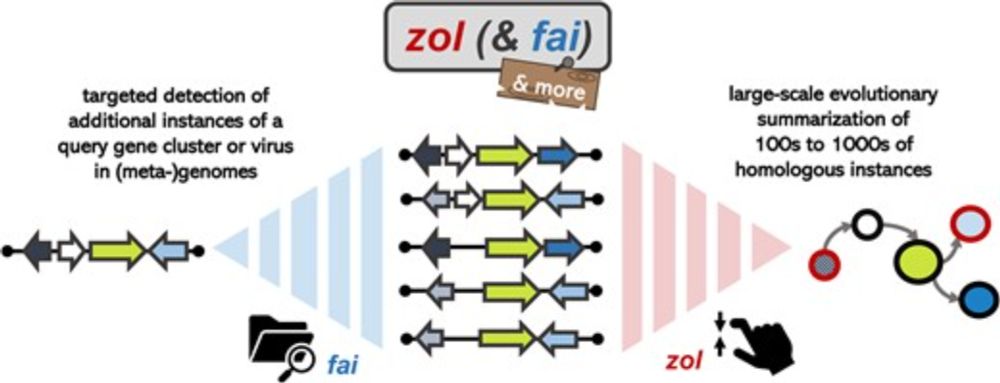
zol and fai: large-scale targeted detection and evolutionary investigation of gene clusters
Abstract. Many universally and conditionally important genes are genomically aggregated within clusters. Here, we introduce fai and zol, which together ena
academic.oup.com
February 5, 2025 at 7:52 PM
Difficult to focus on science during these times, but if you have the bandwidth, our study on zol is now out in NAR: academic.oup.com/nar/article/...
GitHub: github.com/Kalan-Lab/zol
Please give the software a try and let us know if you have any feedback! And stay tuned for new features...
GitHub: github.com/Kalan-Lab/zol
Please give the software a try and let us know if you have any feedback! And stay tuned for new features...

