We found that loss-of-function mutations in the carbapenem entry porin OprD of Pseudomonas aeruginosa do more than confer #AntibioticResistance: they reshape the bacterial membrane and interaction with the host, enhancing epithelial colonization capacity 🦠
#Microsky
We found that loss-of-function mutations in the carbapenem entry porin OprD of Pseudomonas aeruginosa do more than confer #AntibioticResistance: they reshape the bacterial membrane and interaction with the host, enhancing epithelial colonization capacity 🦠
#Microsky
Ever wondered why only a fraction of genomes encode CRISPR immunity? 🧬 🦠
Turns out CRISPR is rarely beneficial against virulent phages, being most beneficial against those for which resistance mutations are rare!
An epic effort by Rosanna Wright
www.biorxiv.org/content/10.1...

Ever wondered why only a fraction of genomes encode CRISPR immunity? 🧬 🦠
Turns out CRISPR is rarely beneficial against virulent phages, being most beneficial against those for which resistance mutations are rare!
An epic effort by Rosanna Wright
www.biorxiv.org/content/10.1...
@rkoszul.bsky.social www.nature.com/articles/s41...

@rkoszul.bsky.social www.nature.com/articles/s41...
elsevier.digitalcommonsdata.com/datasets/btc...
elsevier.digitalcommonsdata.com/datasets/btc...
Plasmids are associated with very variable fitness costs in their different bacterial hosts. But, what is the contribution of each of the plasmid-genes in these host-specific effects? Study led by
@jorgesastred.bsky.social, @sanmillan.bsky.social and myself! 1/14
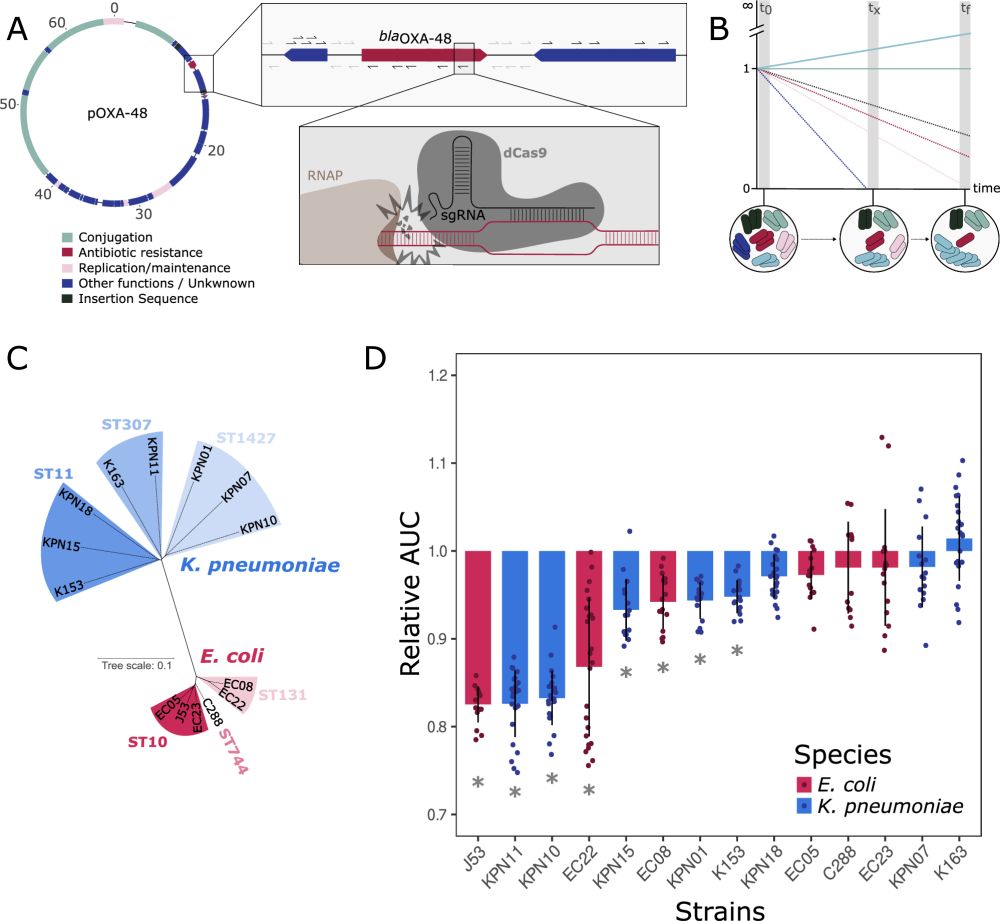
Plasmids are associated with very variable fitness costs in their different bacterial hosts. But, what is the contribution of each of the plasmid-genes in these host-specific effects? Study led by
@jorgesastred.bsky.social, @sanmillan.bsky.social and myself! 1/14
Plasmids promote bacterial evolution through a copy number-driven increase in mutation rate.
We combine theory, simulations, experimental evolution, and bioinformatics to demonstrate that mutation rates scale with plasmid copy number.
Let's dive in! 🧵👇

Plasmids promote bacterial evolution through a copy number-driven increase in mutation rate.
We combine theory, simulations, experimental evolution, and bioinformatics to demonstrate that mutation rates scale with plasmid copy number.
Let's dive in! 🧵👇
In our new paper, we tackled this question using theory, simulations, bioinformatics, and experiments!
👇 Check out all the details in Paula’s thread!
Hint: 🐇 (most of the time)
In our new paper, we tackled this question using theory, simulations, bioinformatics, and experiments!
👇 Check out all the details in Paula’s thread!
Hint: 🐇 (most of the time)
Come for the first large-scale analysis of plasmid copy number across species,
stay for one of the most intriguing results of my lab: universal scaling laws in plasmid biology! 📈🧬
👉 www.nature.com/articles/s41...
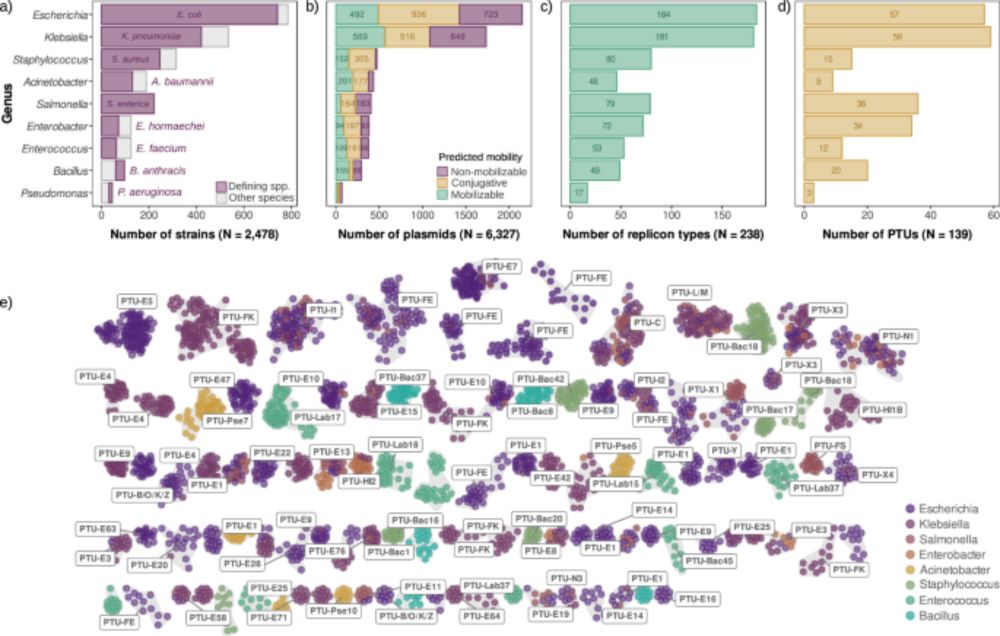

Come for the first large-scale analysis of plasmid copy number across species,
stay for one of the most intriguing results of my lab: universal scaling laws in plasmid biology! 📈🧬
👉 www.nature.com/articles/s41...

Come for the first large-scale analysis of plasmid copy number across species,
stay for one of the most intriguing results of my lab: universal scaling laws in plasmid biology! 📈🧬
👉 www.nature.com/articles/s41...
doi.org/10.1038/s414...
We analyzed thousands of diverse bacterial plasmids to shed light for the first time on a key aspect of plasmid biology: plasmid copy number. 1/7 👇
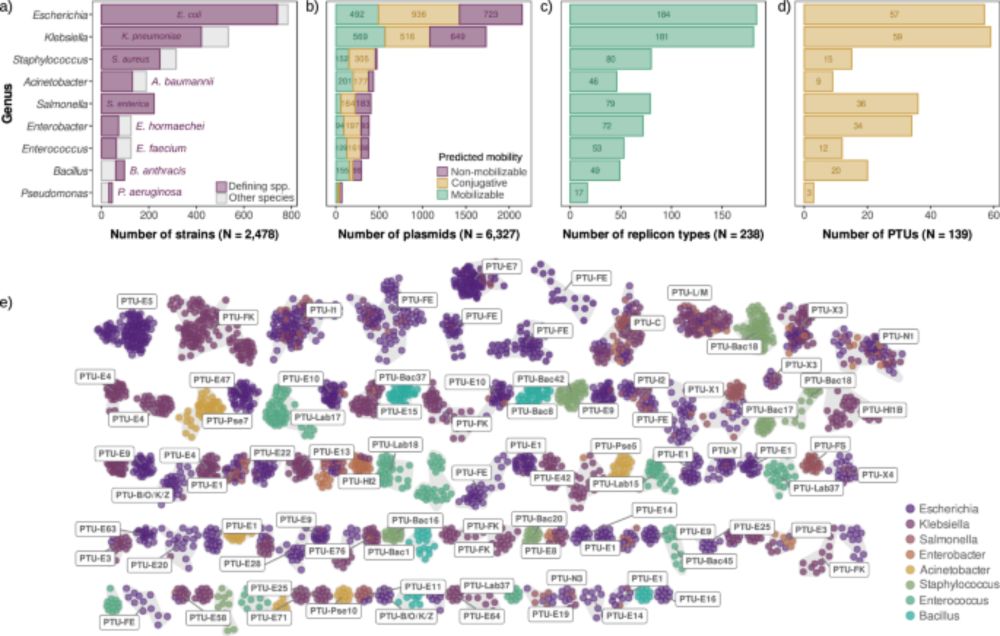
doi.org/10.1038/s414...
We analyzed thousands of diverse bacterial plasmids to shed light for the first time on a key aspect of plasmid biology: plasmid copy number. 1/7 👇



by Laura Toribio-Celestino, @sanmillan.bsky.social et al., in @naturecomms.bsky.social
www.nature.com/articles/s41...
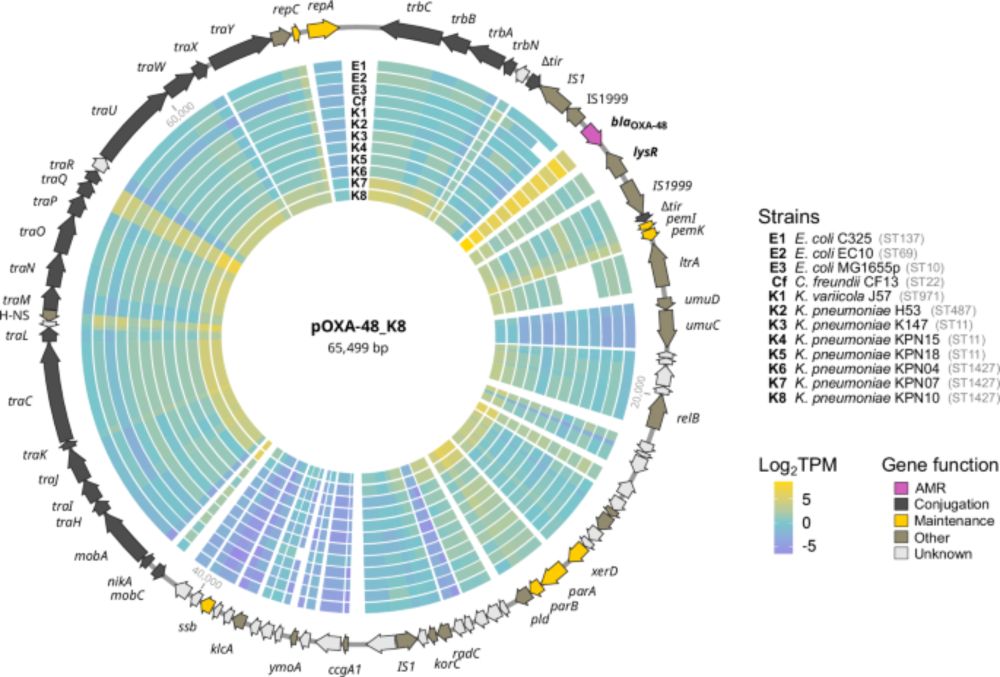
by Laura Toribio-Celestino, @sanmillan.bsky.social et al., in @naturecomms.bsky.social
www.nature.com/articles/s41...

